The conventional approach
Usually when conducting an analysis of variance we follow the steps outlined below:
Step 1. Make model with multiple predictors no interaction.
Step 2. Make another model with the supposed interaction.
Step 3. anova(model1,model2), if the p-value is significant, the more
complex model is the one that you should select.
The tidymodels approach
Let’s take a look at the Palmer Penguins dataset:
Show the code
penguins %>%
head(5) # A tibble: 5 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
# ℹ 2 more variables: sex <fct>, year <int>
We have a few variables of interest which we can hypothesise may be interacting with each other. Let’s visualise the data:
Show the code
penguins %>%
na.omit() %>%
ggplot(aes(bill_length_mm,bill_depth_mm))+
geom_point(aes(color = species)) +
geom_smooth(method = lm, color = "orange",
formula = 'y~x',se=FALSE) +
geom_smooth(method = lm, aes(color = species),
formula = 'y~x',
se=FALSE) +
labs(y="Bill Depth (mm)",
x= "Bill Length (mm)")+
theme_minimal()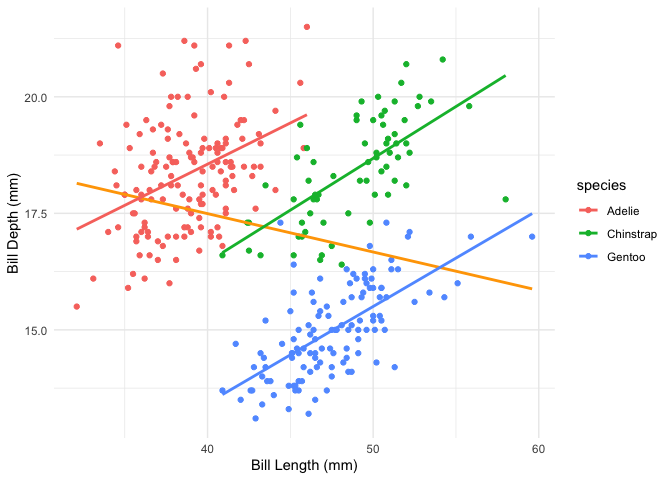
The general linear model doesn’t seem too bad, but it doesn’t take into account potential interactions between species and the variables above. Conventionally we would test this by comparing two models, one which accounts for the interaction and one that doesn’t:
Show the code
model1 <- lm(bill_length_mm ~ bill_depth_mm * species, data = penguins %>% na.omit())
# summary(model1)
model2 <- lm(bill_length_mm ~ bill_depth_mm + species, data = penguins %>% na.omit())
# summary(model2)
anova(model1, model2) %>%
tidy() # A tibble: 2 × 7
term df.residual rss df sumsq statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 bill_length_mm ~ bill_depth_… 327 1954. NA NA NA NA
2 bill_length_mm ~ bill_depth_… 329 2095. -2 -141. 11.8 1.17e-5
We see that the p-value is statistically significant, so we choose the model with the interaction term.
The tidymodels framework
Create the data splits and feature engineering recipes
Let’s now take the tidymodels approach to the same problem. We start by defining our data budget and splits:
Show the code
set.seed(1234)
penguin_split <- initial_split(penguins %>%
na.omit(), strata = species, prop = 0.8)
penguin_train <- training(penguin_split)
penguin_test <- testing(penguin_split)Next, we will define the respective pre-processing recipes, one with the interaction term and another without it:
Show the code
# model without interaction:
rec_normal <-
recipe(bill_length_mm ~ bill_depth_mm + species,
data=penguin_train) %>%
step_center(all_numeric_predictors())
# the model with interaction:
rec_interaction <-
rec_normal %>%
step_interact(~ bill_depth_mm:species)Create the workflows
Now that we have our recipes we can proceed to create our respective workflows:
Show the code
# Define the model
penguin_model <-
linear_reg() %>%
set_engine("lm") %>%
set_mode("regression")
# Workflows
# normal workflow:
penguin_wf <-
workflow() %>%
add_model(penguin_model) %>%
add_recipe(rec_normal)
# interaction workflow
penguin_wf_interaction <-
penguin_wf %>%
update_recipe(rec_interaction)Let’s now fit our model to the data:
Show the code
## Fitting
# use the last_fit() to make sure data is trained on
# training dataset and evaluated on test dataset.
penguin_normal_lf <-
last_fit(penguin_wf,
split = penguin_split)
penguin_inter_lf <-
last_fit(penguin_wf_interaction,
split = penguin_split) → A | warning: Categorical variables used in `step_interact()` should probably be avoided;
This can lead to differences in dummy variable values that are produced by
?step_dummy (`?recipes::step_dummy()`). Please convert all involved variables
to dummy variables first.
There were issues with some computations A: x1
There were issues with some computations A: x1The ANOVA part
Before we compare the models using the anova() function, we have to
extract the fitted models from the respective workflows:
Show the code
normalmodel <- penguin_normal_lf %>% extract_fit_engine()
intermodel <- penguin_inter_lf %>% extract_fit_engine()
anova(normalmodel,intermodel) %>%
tidy() # A tibble: 2 × 7
term df.residual rss df sumsq statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 ..y ~ bill_depth_mm + species 261 1657. NA NA NA NA
2 ..y ~ bill_depth_mm + specie… 259 1533. 2 124. 10.5 4.22e-5
We can get the model metrics using the yardstick package:
Show the code
penguin_inter_lf %>%
collect_metrics() # A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 rmse standard 2.49 pre0_mod0_post0
2 rsq standard 0.782 pre0_mod0_post0
Let’s take a deeper look at the interaction terms:
Show the code
intermodel %>%
tidy() # A tibble: 6 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 37.9 0.310 122. 5.51e-231
2 bill_depth_mm 0.837 0.184 4.55 8.41e- 6
3 speciesChinstrap 8.42 0.590 14.3 1.85e- 34
4 speciesGentoo 14.3 0.663 21.6 5.98e- 60
5 bill_depth_mm_x_speciesChinstrap 1.10 0.343 3.22 1.45e- 3
6 bill_depth_mm_x_speciesGentoo 1.27 0.308 4.11 5.21e- 5
Finally it may be useful to plot the results:
Show the code
# plot model predictions:
penguin_inter_lf %>%
collect_predictions() %>%
ggplot(aes(.pred, bill_length_mm)) +
geom_point() +
geom_abline(intercept = 0, slope = 1, color = "orange", size =1) +
labs(x = "Predicted Bill Length",
y = "Observed Bill Length",
title = "R² plot") +
theme_minimal() Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.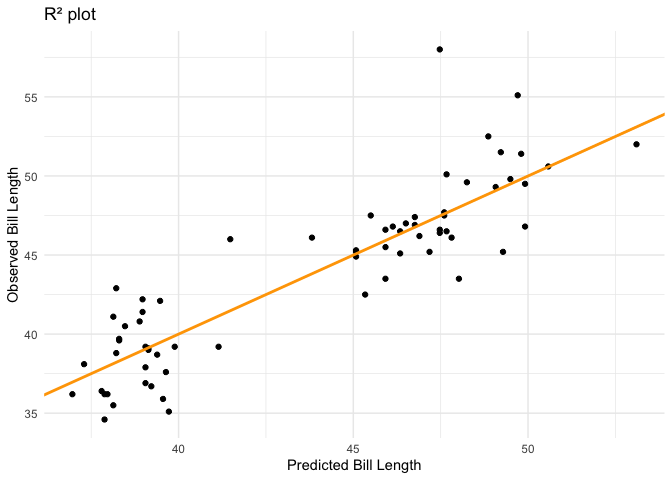
That’s it! We have successfully incorporated the tidymodels framework into the analysis ensuring robustness and sticking to good data practices.